Here are the classes, structs, unions and interfaces with brief descriptions:
[detail level 12345]
| ►Nmadness | Holds machinery to set up Functions/FuncImpls using various Factories and Interfaces |
| ►Narchive | |
| Carchive_array | Wrapper for dynamic arrays and pointers |
| Carchive_ptr | Wrapper for opaque pointer ... bitwise copy of the pointer ... no remapping performed |
| Carchive_typeinfo | |
| CArchiveImpl | Default implementation of wrap_store and wrap_load |
| CArchiveImpl< Archive, archive_array< T > > | Partial specialization for archive_array |
| CArchiveImpl< Archive, T[n]> | Partial specialization for fixed dimension array redirects to archive_array |
| CArchiveImpl< ParallelInputArchive, archive_array< T > > | Read archive array and broadcast |
| CArchiveImpl< ParallelInputArchive, T > | |
| CArchiveImpl< ParallelInputArchive, T[n]> | Forward fixed size array to archive_array |
| CArchiveImpl< ParallelOutputArchive, archive_array< T > > | Write archive array only from process zero |
| CArchiveImpl< ParallelOutputArchive, T > | |
| CArchiveImpl< ParallelOutputArchive, T[n]> | Forward fixed size array to archive_array |
| CArchiveLoadImpl | Default load of a thingy via serialize(ar,t) |
| CArchiveLoadImpl< Archive, const DerivativeBase< T, NDIM > * > | |
| CArchiveLoadImpl< Archive, const FunctionImpl< T, NDIM > * > | |
| CArchiveLoadImpl< Archive, const SeparatedConvolution< T, NDIM > * > | |
| CArchiveLoadImpl< Archive, detail::RemoteCounter > | |
| CArchiveLoadImpl< Archive, detail::WorldPtr< T > > | |
| CArchiveLoadImpl< Archive, FunctionImpl< T, NDIM > * > | |
| CArchiveLoadImpl< Archive, Future< T > > | Deserialize a future into an unassigned future |
| CArchiveLoadImpl< Archive, Future< void > > | Deserialize a future into an unassigned future |
| CArchiveLoadImpl< Archive, Future< Void > > | Deserialize a future into an unassigned future |
| CArchiveLoadImpl< Archive, GenTensor< T > > | Deserialize a tensor ... existing tensor is replaced |
| CArchiveLoadImpl< Archive, Key< NDIM > > | |
| CArchiveLoadImpl< Archive, std::array< T, N > > | |
| CArchiveLoadImpl< Archive, std::complex< T > > | Deserialize a complex number |
| CArchiveLoadImpl< Archive, std::map< T, Q > > | Deserialize an STL map. Map is NOT cleared; duplicate elements are replaced |
| CArchiveLoadImpl< Archive, std::shared_ptr< const FunctionImpl< T, NDIM > > > | |
| CArchiveLoadImpl< Archive, std::shared_ptr< FunctionImpl< T, NDIM > > > | |
| CArchiveLoadImpl< Archive, std::string > | Deserialize STL string. Clears & resizes as necessary |
| CArchiveLoadImpl< Archive, std::vector< bool > > | Deserialize STL vector<bool>. Clears & resizes as necessary |
| CArchiveLoadImpl< Archive, std::vector< Future< T > > > | Deserialize a future into an unassigned future |
| CArchiveLoadImpl< Archive, std::vector< T > > | Deserialize STL vector. Clears & resizes as necessary |
| CArchiveLoadImpl< Archive, Tensor< T > > | Deserialize a tensor ... existing tensor is replaced |
| CArchiveLoadImpl< Archive, World * > | |
| CArchiveLoadImpl< BinaryFstreamInputArchive, Key< NDIM > > | |
| CArchiveLoadImpl< BufferInputArchive, const WorldObject< Derived > * > | |
| CArchiveLoadImpl< BufferInputArchive, WorldObject< Derived > * > | |
| CArchiveLoadImpl< ParallelInputArchive, Function< T, NDIM > > | |
| CArchiveLoadImpl< ParallelInputArchive, WorldContainer< keyT, valueT > > | |
| CArchivePrePostImpl | Default implementation of pre/postamble |
| CArchivePrePostImpl< BufferInputArchive, T > | |
| CArchivePrePostImpl< BufferOutputArchive, T > | |
| CArchivePrePostImpl< MPIInputArchive, T > | |
| CArchivePrePostImpl< MPIOutputArchive, T > | |
| CArchivePrePostImpl< MPIRawInputArchive, T > | |
| CArchivePrePostImpl< MPIRawOutputArchive, T > | |
| CArchivePrePostImpl< ParallelInputArchive, T > | Disable type info for parallel input archives |
| CArchivePrePostImpl< ParallelOutputArchive, T > | Disable type info for parallel output archives |
| CArchivePrePostImpl< TextFstreamInputArchive, T > | |
| CArchivePrePostImpl< TextFstreamOutputArchive, T > | |
| CArchiveSerializeImpl | Default symmetric serialization of a non-fundamental thingy |
| CArchiveSerializeImpl< Archive, std::pair< T, Q > > | (de)Serialize an STL pair |
| CArchiveStoreImpl | Default store of a thingy via serialize(ar,t) |
| CArchiveStoreImpl< Archive, const DerivativeBase< T, NDIM > * > | |
| CArchiveStoreImpl< Archive, const FunctionImpl< T, NDIM > * > | |
| CArchiveStoreImpl< Archive, const SeparatedConvolution< T, NDIM > * > | |
| CArchiveStoreImpl< Archive, detail::RemoteCounter > | |
| CArchiveStoreImpl< Archive, detail::WorldPtr< T > > | |
| CArchiveStoreImpl< Archive, FunctionImpl< T, NDIM > * > | |
| CArchiveStoreImpl< Archive, Future< T > > | Serialize an assigned future |
| CArchiveStoreImpl< Archive, Future< void > > | Serialize an assigned future |
| CArchiveStoreImpl< Archive, Future< Void > > | Serialize an assigned future |
| CArchiveStoreImpl< Archive, GenTensor< T > > | Serialize a tensor |
| CArchiveStoreImpl< Archive, Key< NDIM > > | |
| CArchiveStoreImpl< Archive, std::array< T, N > > | |
| CArchiveStoreImpl< Archive, std::complex< T > > | Serialize a complex number |
| CArchiveStoreImpl< Archive, std::map< T, Q > > | Serialize an STL map (crudely) |
| CArchiveStoreImpl< Archive, std::shared_ptr< const FunctionImpl< T, NDIM > > > | |
| CArchiveStoreImpl< Archive, std::shared_ptr< FunctionImpl< T, NDIM > > > | |
| CArchiveStoreImpl< Archive, std::string > | Serialize STL string |
| CArchiveStoreImpl< Archive, std::vector< bool > > | Serialize STL vector<bool> (as plain array of bool) |
| CArchiveStoreImpl< Archive, std::vector< Future< T > > > | |
| CArchiveStoreImpl< Archive, std::vector< T > > | Serialize STL vector |
| CArchiveStoreImpl< Archive, Tensor< T > > | Serialize a tensor |
| CArchiveStoreImpl< Archive, World * > | |
| CArchiveStoreImpl< BufferOutputArchive, const WorldObject< Derived > * > | |
| CArchiveStoreImpl< BufferOutputArchive, WorldObject< Derived > * > | |
| CArchiveStoreImpl< ParallelOutputArchive, Function< T, NDIM > > | |
| CArchiveStoreImpl< ParallelOutputArchive, WorldContainer< keyT, valueT > > | Write container to parallel archive with optional fence |
| CBaseArchive | Base class for all archives classes |
| CBaseInputArchive | Base class for input archives classes |
| CBaseOutputArchive | Base class for output archives classes |
| CBaseParallelArchive | Base class for input and output parallel archives |
| CBinaryFstreamInputArchive | Wraps an archive around a binary file stream for input |
| CBinaryFstreamOutputArchive | Wraps an archive around a binary file stream for output |
| CBufferInputArchive | Wraps an archive around a memory buffer for input |
| CBufferOutputArchive | Wraps an archive around a memory buffer for output |
| Cis_archive | Checks that T is an archive type |
| Cis_input_archive | Checks that T is an input archive type |
| Cis_output_archive | Checks that T is an output archive type |
| CMPIInputArchive | |
| CMPIOutputArchive | |
| CMPIRawInputArchive | |
| CMPIRawOutputArchive | |
| CParallelInputArchive | An archive for storing local or parallel data wrapping BinaryFstreamInputArchive |
| CParallelOutputArchive | An archive for storing local or parallel data wrapping BinaryFstreamOutputArchive |
| CParallelSerializableObject | Objects that implement their own parallel archive interface should derive from this |
| CTextFstreamInputArchive | Wraps an archive around a text file stream for input |
| CTextFstreamOutputArchive | Wraps an archive around a text file stream for output |
| CVectorInputArchive | Wraps an archive around an STL vector for input |
| CVectorOutputArchive | Wraps an archive around an STL vector for output |
| ►Ndetail | |
| Cabsinplace | |
| Cabssqop | |
| Cabssquareinplace | |
| Cadqtest | |
| CArgCount | |
| CArgCountHelper | |
| CArgCountHelper< void > | |
| CArgHolder | A wrapper object for holding task function objects |
| CCheckedArrayDeleter | Checked array pointer delete functor |
| CCheckedDeleter | Checked pointer delete functor |
| CCheckedFree | Deleter to free memory for a shared_ptr using free() |
| CDefaultInitPtr | |
| CDefaultInitPtr< std::shared_ptr< T > > | |
| CDeferredCleanup | Deferred cleanup of shared_ptr's |
| CDistCache | Distributed caching utility |
| CForEachRootTask | Apply an operation to a range of iterators |
| CForEachTask | Apply an operation to a range of iterators |
| Cfunction_enabler | |
| Chelper | |
| Chelper< T, 0 > | |
| Chelper< T, 1 > | |
| Cimagop | |
| Cinfo | |
| Cinfo_base | |
| Cis_movable | Type trait for movable objects |
| Cis_movable< MoveWrapper< T > > | Type trait for movable objects |
| Cmemfunc_enabler | |
| CMemFuncWrapper | |
| CMemFuncWrapper< ptrT, memfnT, void > | |
| CMoveWrapper | Wrapper for movable objects |
| CNoDeleter | Use this deleter with shared_ptr to do nothing for the pointer cleanup |
| Cnoop | |
| CPendingMsg | |
| Cptr_traits | |
| Cptr_traits< void > | |
| Crealop | |
| CReferenceWrapper | Reference wrapper class |
| CRemoteCounter | Remote reference counter |
| CRemoteCounterBase | Base class for remote counter implementation objects |
| CRemoteCounterImpl | Remote counter implementation object |
| Cscaleinplace | |
| Csquareinplace | |
| Ctask_arg | |
| Ctask_arg< Future< T > > | |
| Ctask_arg< Future< void > > | |
| Ctask_arg< void > | |
| Ctask_result_type | |
| CTaskHandlerInfo | |
| CWorldMpi | MPI singleton that manages MPI setup and teardown for MADNESS |
| CWorldMpiRuntime | MPI runtime reference counter |
| CWorldPtr | A global pointer address, valid anywhere in the world |
| ►NHash_private | |
| Cbin | |
| Centry | |
| CHashAccessor | |
| CHashIterator | Iterator for hash |
| ►Ntr1 | |
| ►Narray | |
| Carray | Array idential to C++0X arrays |
| Carray< T, 0 > | |
| ►Ndetail | |
| Cfunction_traits | Function traits in the spirt of boost function traits |
| Cfunction_traits< returnT(*)()> | Function traits in the spirt of boost function traits |
| Cfunction_traits< returnT(*)(arg1T)> | Function traits in the spirt of boost function traits |
| Cfunction_traits< returnT(*)(arg1T, arg2T)> | Function traits in the spirt of boost function traits |
| Cfunction_traits< returnT(*)(arg1T, arg2T, arg3T)> | Function traits in the spirt of boost function traits |
| Cfunction_traits< returnT(*)(arg1T, arg2T, arg3T, arg4T)> | Function traits in the spirt of boost function traits |
| Cfunction_traits< returnT(*)(arg1T, arg2T, arg3T, arg4T, arg5T)> | Function traits in the spirt of boost function traits |
| Cfunction_traits< returnT(*)(arg1T, arg2T, arg3T, arg4T, arg5T, arg6T)> | Function traits in the spirt of boost function traits |
| Cfunction_traits< returnT(*)(arg1T, arg2T, arg3T, arg4T, arg5T, arg6T, arg7T)> | Function traits in the spirt of boost function traits |
| Cfunction_traits< returnT(*)(arg1T, arg2T, arg3T, arg4T, arg5T, arg6T, arg7T, arg8T)> | Function traits in the spirt of boost function traits |
| Cfunction_traits< returnT(*)(arg1T, arg2T, arg3T, arg4T, arg5T, arg6T, arg7T, arg8T, arg9T)> | Function traits in the spirt of boost function traits |
| CHost | |
| Cmemfunc_traits | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)() const > | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)()> | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)(arg1T) const > | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)(arg1T)> | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)(arg1T, arg2T) const > | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)(arg1T, arg2T)> | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)(arg1T, arg2T, arg3T) const > | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)(arg1T, arg2T, arg3T)> | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)(arg1T, arg2T, arg3T, arg4T) const > | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)(arg1T, arg2T, arg3T, arg4T)> | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)(arg1T, arg2T, arg3T, arg4T, arg5T) const > | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)(arg1T, arg2T, arg3T, arg4T, arg5T)> | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)(arg1T, arg2T, arg3T, arg4T, arg5T, arg6T) const > | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)(arg1T, arg2T, arg3T, arg4T, arg5T, arg6T)> | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)(arg1T, arg2T, arg3T, arg4T, arg5T, arg6T, arg7T) const > | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)(arg1T, arg2T, arg3T, arg4T, arg5T, arg6T, arg7T)> | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)(arg1T, arg2T, arg3T, arg4T, arg5T, arg6T, arg7T, arg8T) const > | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)(arg1T, arg2T, arg3T, arg4T, arg5T, arg6T, arg7T, arg8T)> | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)(arg1T, arg2T, arg3T, arg4T, arg5T, arg6T, arg7T, arg8T, arg9T) const > | Member function traits in the spirt of boost function traits |
| Cmemfunc_traits< returnT(objT::*)(arg1T, arg2T, arg3T, arg4T, arg5T, arg6T, arg7T, arg8T, arg9T)> | Member function traits in the spirt of boost function traits |
| Cresult_of | |
| Cresult_of< fnT, typename enable_if_c< function_traits< fnT >::value >::type > | |
| Cresult_of< fnT, typename enable_if_c< memfunc_traits< fnT >::value >::type > | |
| ►Nshptr | |
| ►Ndetail | |
| Cconst_cast_tag | |
| Cdynamic_cast_tag | |
| Cenable_if_convertible | |
| Cenable_if_convertible_impl | |
| Cenable_if_convertible_impl< false > | |
| Cenable_if_convertible_impl< true > | |
| Cesft2_deleter_wrapper | |
| Cshared_count | |
| Cshared_ptr_traits | |
| Cshared_ptr_traits< void > | |
| Cshared_ptr_traits< void const > | |
| Cshared_ptr_traits< void const volatile > | |
| Cshared_ptr_traits< void volatile > | |
| Csp_convertible | |
| Csp_counted_base | |
| Csp_counted_impl_p | |
| Csp_counted_impl_pd | |
| Csp_empty | |
| Csp_nothrow_tag | |
| Cstatic_cast_tag | |
| Cweak_count | |
| Cbad_weak_ptr | |
| Cenable_shared_from_this | |
| Cenable_shared_from_this2 | |
| Cshared_ptr | |
| Cweak_ptr | |
| Cadd_const | Add_const<T>::type will be const T |
| Cadd_const< const T > | Add_const<T>::type will be const T |
| Cenable_if | Std::enable_if is similar to boost::enable_if_c for conditionally instantiating templates based on type |
| Cenable_if< false, returnT > | Std::enable_if is similar to boost::enable_if_c for conditionally instantiating templates based on type |
| Cintegral_constant | |
| Cis_arithmetic | |
| Cis_array | Is_array<T>::value will be true if T is an array type |
| Cis_array< T[n]> | Is_array<T>::value will be true if T is an array type |
| Cis_base_of | |
| Cis_compound | |
| Cis_const | Is_const<T>::value will be true if T is const |
| Cis_const< const T > | Is_const<T>::value will be true if T is const |
| Cis_floating_point | Is_floating_point<T>::value will be true if T is a fundamental floating-point type |
| Cis_function | Is_function<T>::value is true if T is a function pointer |
| Cis_fundamental | Is_fundamental<T>::value will be true if T is a fundamental type |
| Cis_integral | Is_integral<T>::value will be true if T is a fundamental integer type |
| Cis_member_function_pointer | Is_member_function_pointer<T>::value is true if T is a member function pointer |
| Cis_object | |
| Cis_pointer | Is_pointer<T>::value will be true if T is a pointer type |
| Cis_pointer< T * > | Is_pointer<T>::value will be true if T is a pointer type |
| Cis_reference | Is_reference<T>::value will be true if T is a reference type |
| Cis_reference< T & > | Is_reference<T>::value will be true if T is a reference type |
| Cis_same | Is_same<A,B> returns true if A and B are the same type |
| Cis_same< A, A > | Is_same<A,B> returns true if A and B are the same type |
| Cis_void | True if T is void |
| Cis_void< void > | True if T is void |
| Cremove_const | Remove_const<const T>::type will be T |
| Cremove_const< const T > | Remove_const<const T>::type will be T |
| Cremove_cv | Remove_cv<const T>::type or remove_cv<volatile T>::type will be T |
| Cremove_pointer | Remove_pointer<T*>::type will be T |
| Cremove_pointer< T * > | Remove_pointer<T>::type will be T |
| Cremove_reference | Remove_reference<&T>::type will be T |
| Cremove_reference< T & > | Remove_reference<&T>::type will be T |
| Cremove_volatile | Remove_volatile<volatile T>::type will be T |
| Cremove_volatile< volatile T > | Remove_volatile<volatile T>::type will be T |
| Cabs_op | |
| Cabs_square_op | |
| CAbstractVectorSpace | A generic vector space which provides common operations needed by linear algebra routines (norm, inner product, etc.) |
| Callocator | |
| CAmArg | World active message that extends an RMI message |
| Capply_kernel_functor | |
| CAtom | |
| CAtomCore | |
| CAtomicBasis | Represents multiple shells of contracted gaussians on a single center |
| CAtomicBasisFunction | Used to represent one basis function from a shell on a specific center |
| CAtomicBasisFunctor | |
| ►CAtomicBasisSet | Contracted Gaussian basis |
| CAnalysisSorter | |
| CAtomicData | |
| CAtomicInt | An integer with atomic set, get, read+inc, read+dec, dec+test operations |
| CBarrier | |
| CBaseTensor | The base class for tensors defines generic capabilities |
| CBindNullaryConstMemFun | Simple binder for const member functions with no arguments |
| CBindNullaryConstMemFun< T, void > | Specialization of BindNullaryConstMemFun for void return |
| CBindNullaryMemFun | Simple binder for member functions with no arguments |
| CBindNullaryMemFun< T, void > | Specialization of BindNullaryMemFun for void return |
| CBinSorter | A parallel bin sort across MPI processes |
| CBoundaryConditions | This class is used to specify boundary conditions for all operatorsExterior boundary conditions (i.e., on the simulation domain) are associated with operators (not functions). The types of boundary conditions available are in the enum BCType |
| CBSHFunctionInterface | Function like f(x) = exp(-mu x)/x |
| CCalculationParameters | |
| CCallbackInterface | This class used for callbacks (e.g., for dependency tracking) |
| CCIS | Holds all machinery to compute excited state properties |
| CCoeffTracker | Class to track where relevant (parent) coeffs are |
| CComplexExp | |
| CCompositeFactory | Factory for facile setup of a CompositeFunctorInterface and its FuncImpl |
| CCompositeFunctorInterface | CompositeFunctorInterface implements a wrapper of holding several functions and functors |
| CConcurrentHashMap | |
| Cconditional_conj_struct | For real types return value, for complex return conjugate |
| Cconditional_conj_struct< Q, true > | For real types return value, for complex return conjugate |
| CConditionVariable | Scalable and fair condition variable (spins on local value) |
| Cconj_op | |
| CContractedGaussianShell | Represents a single shell of contracted, Cartesian, Gaussian primitives |
| CConvolution1D | Provides the common functionality/interface of all 1D convolutions |
| CConvolutionData1D | !!! Note that if Rnormf is zero then ***ALL*** of the tensors are empty |
| CConvolutionND | Array of 1D convolutions (one / dimension) |
| CCoreOrbital | |
| CCoreOrbitalDerivativeFunctor | |
| CCoreOrbitalFunctor | |
| CCorePotential | Represents a core potential |
| CCorePotentialDerivativeFunctor | |
| CCorePotentialManager | |
| Cdefault_allocator | |
| CDeferredDeleter | Deferred deleter for smart pointers |
| CDependencyInterface | Provides interface for tracking dependencies |
| CDerivative | Implements derivatives operators with variety of boundary conditions on simulation domain |
| CDerivativeBase | Tri-diagonal operator traversing tree primarily for derivative operator |
| CDFT | |
| CDFTCoulombOp | |
| CDFTCoulombPeriodicOp | |
| CDFTNuclearChargeDensityOp | |
| CDFTNuclearPotentialOp | |
| Cdiffuse_functions | Functor that adds diffuse 1s functions on given coordinates with given signs (phases) |
| CDipoleFunctor | A MADNESS functor to compute either x, y, or z |
| Cdisable_if | Disable_if from Boost for conditionally instantiating templates based on type |
| Cdisable_if_c | Disable_if from Boost for conditionally instantiating templates based on type |
| Cdisable_if_c< true, returnT > | Disable_if from Boost for conditionally instantiating templates based on type |
| CDisplacements | Holds displacements for applying operators to avoid replicating for all operators |
| CDistributedMatrix | Manages data associated with a row/column/block distributed array |
| CDistributedMatrixDistribution | |
| CDomainMaskInterface | The interface for masking functions defined by signed distance functions |
| CDomainMaskSDFFunctor | Framework for combining a signed distance function (sdf) with a domain mask to produce MADNESS functions |
| CDQStats | |
| CDQueue | A thread safe, fast but simple doubled-ended queue |
| CEigSolver | |
| CEigSolverOp | |
| CElectronicStructureParams | |
| CElectronPair | Enhanced POD for the pair functions |
| CElectronRepulsionInterface | Function like f(x)=1/x |
| CElementaryInterface | ElementaryInterface (formerly FunctorInterfaceWrapper) interfaces a c-function |
| Cenable_if | Enable_if from Boost for conditionally instantiating templates based on type |
| Cenable_if_c | Enable_if_c from Boost for conditionally instantiating templates based on type |
| Cenable_if_c< false, returnT > | Enable_if_c from Boost for conditionally instantiating templates based on type |
| Cerror_leaf_op | Returns true if the node is well represented compared to its parent |
| Cexoperators | |
| CFGFactory | Factory to set up an ElectronRepulsion Function |
| CFGInterface | Function like f(x) = (1 - exp(-mu x))/x |
| ►CFunction | A multiresolution adaptive numerical function |
| Cautorefine_square_op | |
| CSimpleUnaryOpWrapper | |
| Cfunction_real2complex_op | |
| CFunctionCommonData | FunctionCommonData holds all Function data common for given k |
| CFunctionDefaults | FunctionDefaults holds default paramaters as static class members |
| CFunctionFactory | FunctionFactory implements the named-parameter idiom for Function |
| CFunctionFunctorInterface | Abstract base class interface required for functors used as input to Functions |
| ►CFunctionImpl | FunctionImpl holds all Function state to facilitate shallow copy semantics |
| Cadd_op | Add two functions f and g: result=alpha * f + beta * g |
| Ccoeff_value_adaptor | |
| Cdo_average | "put" this on g |
| Cdo_change_tensor_type | Change representation of nodes' coeffs to low rank, optional fence |
| Cdo_check_symmetry_local | Check symmetry wrt particle exchange |
| Cdo_consolidate_buffer | |
| Cdo_convert_to_color | |
| Cdo_err_box | |
| Cdo_gaxpy_inplace | |
| Cdo_inner_ext_local | |
| Cdo_inner_ext_local_ffi | |
| Cdo_inner_local | Compute the inner product of this range with other |
| Cdo_keep_sum_coeffs | Keep only the sum coefficients in each node |
| Cdo_mapdim | Map this on f |
| Cdo_merge_trees | Merge the coefficent boxes of this into other's tree |
| Cdo_norm2sq_local | |
| Cdo_op_args | Laziness |
| Cdo_reduce_rank | Reduce the rank of the nodes, optional fence |
| Cdo_standard | Changes non-standard compressed form to standard compressed form |
| Cdo_truncate_NS_leafs | Given an NS tree resulting from a convolution, truncate leafs if appropriate |
| Cdo_unary_op_value_inplace | |
| Chartree_op | Hartree product of two LDIM functions to yield a NDIM = 2*LDIM function |
| Cmultiply_op | Perform this multiplication: h(1,2) = f(1,2) * g(1) |
| Cproject_out_op | Project the low-dim function g on the hi-dim function f: result(x) = <f(x,y) | g(y)> |
| Crecursive_apply_op | Recursive part of recursive_apply |
| Crecursive_apply_op2 | Recursive part of recursive_apply |
| Cremove_internal_coeffs | |
| Ctrue_refine_test | |
| CVphi_op_NS | Given a ket and the 1- and 2-electron potentials, construct the function V phi |
| CFunctionInterface | FunctionInterface implements a wrapper around any class with the operator()() |
| CFunctionNode | FunctionNode holds the coefficients, etc., at each node of the 2^NDIM-tree |
| CFunctionSpace | A vector space using MADNESS Functions |
| CFunctorInterface | FunctorInterface interfaces a class or struct with an operator()() |
| CFuture | A future is a possibly yet unevaluated value |
| CFuture< detail::ReferenceWrapper< T > > | Future for holding ReferenceWrapper objects |
| CFuture< Future< T > > | A future of a future is forbidden (by private constructor) |
| CFuture< std::vector< Future< T > > > | Specialization of Future for vector of Futures |
| CFuture< void > | Specialization of Future<void> for internal convenience ... does nothing useful! |
| CFuture< Void > | Specialization of Future<Void> for internal convenience ... does nothing useful! |
| CFutureImpl | Implements the functionality of Futures |
| CFutureImpl< void > | Specialization of FutureImpl<void> for internal convenience ... does nothing useful! |
| CFutureImpl< Void > | Specialization of FutureImpl<Void> for internal convenience ... does nothing useful! |
| CGaussianConvolution1D | 1D convolution with (derivative) Gaussian; coeff and expnt given in simulation coordinates [0,1] |
| CGaussianConvolution1DCache | |
| CGaussianDomainMask | Use a Gaussian for the surface function and the corresponding erf for the domain mask |
| CGaussianGenericFunctor | |
| ►CGenericConvolution1D | Generic 1D convolution using brute force (i.e., slow) adaptive quadrature for rnlp |
| CShmoo | |
| CGenTensor | |
| Cget_derivatives | |
| CGFit | |
| CGroup | A collection of processes |
| CGTHPseudopotential | |
| Chartree_convolute_leaf_op | |
| Chartree_leaf_op | Returns true if the result of a hartree_product is a leaf node (compute norm & error) |
| CHartreeFock | |
| CHartreeFockCoulombOp | |
| CHartreeFockExchangeOp | |
| CHartreeFockNuclearPotentialOp | |
| Chas_result_type | |
| CHash | Hash functor |
| CHighDimIndexIterator | |
| CIEigSolverObserver | |
| Cif_ | |
| Cif_c | Enable_if_same (from Boost?) for conditionally instantiating templates if two types are equal |
| Cif_c< false, T1, T2 > | |
| Cimag_op | |
| CIndexIterator | |
| Cinsert_op | |
| Cis_derived_from | True if A is derived from B and is not B |
| Cis_derived_from< A, A > | True if A is derived from B and is not B |
| Cis_future | Boost-type-trait-like testing of if a type is a future |
| Cis_future< Future< T > > | Boost-type-trait-like testing of if a type is a future |
| Cis_reference_wrapper | Type trait for reference wrapper |
| Cis_reference_wrapper< detail::ReferenceWrapper< T > > | |
| Cis_reference_wrapper< detail::ReferenceWrapper< T > const > | |
| Cis_reference_wrapper< detail::ReferenceWrapper< T > const volatile > | |
| Cis_reference_wrapper< detail::ReferenceWrapper< T > volatile > | |
| Cis_serializable | This defines stuff that is serialiable by default rules ... basically anything contiguous |
| CIsSupported | |
| CIsSupported< TypeData, ReturnType, true > | |
| Ckain_solver_helper_struct | The structure needed if the kain solver shall be used |
| CKey | Key is the index for a node of the 2^NDIM-tree |
| CKeyChildIterator | Iterates in lexical order thru all children of a key |
| CKPoint | |
| Clazy_disable_if | Lazy_disable_if from Boost for conditionally instantiating templates based on type |
| Clazy_disable_if_c | Lazy_disable_if_c from Boost for conditionally instantiating templates based on type |
| Clazy_disable_if_c< true, returnT > | Lazy_disable_if_c from Boost for conditionally instantiating templates based on type |
| Clazy_enable_if | Lazy_enable_if from Boost for conditionally instantiating templates based on type |
| Clazy_enable_if_c | Lazy_enable_if_c from Boost for conditionally instantiating templates based on type |
| Clazy_enable_if_c< false, returnT > | Lazy_enable_if_c from Boost for conditionally instantiating templates based on type |
| CLBCost | |
| Clbcost | |
| CLBDeuxPmap | |
| CLBNodeDeux | |
| Cleaf_op | Returns true if the function has a leaf node at key (works only locally) |
| CLevelPmap | A pmap that locates children on odd levels with their even level parents |
| CLLRVDomainMask | Provides the Li-Lowengrub-Ratz-Voight (LLRV) domain mask characteristic functions |
| ►CLoadBalanceDeux | |
| CCostPerProc | |
| CLoadBalImpl | |
| CLocalizeBoys | |
| CLowDimIndexIterator | |
| CMadnessException | Most exceptions thrown in MADNESS should be derived from these |
| Cmake_fxc | |
| CMatrixInnerTask | |
| CMolecularCorePotentialFunctor | |
| CMolecularDerivativeFunctor | |
| CMolecularEnergy | |
| CMolecularGuessDensityFunctor | |
| CMolecularPotentialFunctor | |
| CMolecule | |
| CMomentFunctor | A MADNESS functor to compute the cartesian moment x^i * y^j * z^k (i, j, k integer and >= 0) |
| CMP2 | Class for computing the first order wave function and MP2 pair energies |
| Cmul_leaf_op | |
| CMutex | Mutex using pthread mutex operations |
| CMutexFair | A scalable and fair mutex (not recursive) |
| CMutexReaderWriter | |
| CMutexWaiter | |
| CMyPmap | Procmap implemented using Tree of TreeCoords |
| CNemo | The Nemo class |
| CNonlinearSolverND | A simple Krylov-subspace nonlinear equation solver |
| CNonstandardIndexIterator | |
| Cnoop | |
| Cop_leaf_op | |
| COperator | A generic operator: takes in one T and produces another T |
| COptimizationTargetInterface | The interface to be provided by functions to be optimized |
| COptimizerInterface | The interface to be provided by optimizers |
| Cperturbed_vxc | |
| CPoolTaskInterface | Lowest level task interface |
| CPoolTaskNull | A no-op task used for various purposes |
| CPotentialManager | |
| CProcessKey | Key object that included the process information |
| CProfileStat | Simple container for parallel profile statistic |
| CProjector | Simple projector class for 1- and 2-particle projectors |
| CProjRLMFunctor | |
| CProjRLMStore | |
| CPthreadConditionVariable | Simple wrapper for Pthread condition variable with its own mutex |
| Cqmsg | |
| CQuasiNewton | Optimization via quasi-Newton (BFGS or SR1 update) |
| CRandom | A random number generator (portable, vectorized, and thread-safe) |
| CRandomState | |
| CRange | Range vaguely a la Intel TBB encapsulates random-access STL-like start and end iterators with chunksize |
| Creal_op | |
| CRecursiveMutex | Recursive mutex using pthread mutex operations |
| CReference | Used to provide rvalue references to support move semantics |
| CRemoteReference | Simple structure used to manage references/pointers to remote instances |
| Cremove_fcvr | |
| Cremove_future | Boost-type-trait-like mapping of Future<T> to T |
| Cremove_future< Future< T > > | Boost-type-trait-like mapping of Future<T> to T |
| CReturnWrapper | Wrapper so that can return something even if returning void |
| CReturnWrapper< void > | Wrapper so that can return something even if returning void |
| CRMI | |
| CRMIStats | |
| CSCF | |
| CScopedArray | Scoped array |
| CScopedMutex | Mutex that is applied/released at start/end of a scope |
| CScopedPtr | Scoped pointer |
| CSDFBox | A box (3 dimensions) |
| CSDFCircle | A circle (2 dimensions) |
| CSDFCone | A cone (3 dimensions) |
| CSDFCube | A cube (3 dimensions) |
| CSDFCylinder | A cylinder (3 dimensions) |
| CSDFEllipsoid | An ellipsoid (3 dimensions) |
| CSDFParaboloid | A paraboloid (3 dimensions) |
| CSDFPlane | A plane surface (3 dimensions) |
| CSDFRectangle | A rectangle (2 dimensions) |
| CSDFSphere | A spherical surface (3 dimensions) |
| CSeparatedConvolution | Convolutions in separated form (including Gaussian) |
| CSeparatedConvolutionData | SeparatedConvolutionData keeps data for all terms, all dimensions |
| CSeparatedConvolutionInternal | |
| CShallowNode | Shallow-copy, pared-down version of FunctionNode, for special purpose only |
| CSignedDFInterface | The interface for a signed distance function (sdf) |
| CSimpleCache | Simplified interface around hash_map to cache stuff for 1D |
| CSimplePmap | A simple process map |
| CSlaterF12Interface | Function like f(x) = (1 - exp(-mu x))/(2 gamma) |
| CSlaterFunctionInterface | Function like f(x)=exp(-mu x) |
| CSlice | A slice defines a sub-range or patch of a dimension |
| CSliceGenTensor | |
| ►CSolver | The main class of the periodic DFT solver
|
| CGuessDensity | |
| CSolverInterface | The interface to be provided by solvers ... NOT USED ANYWHERE? |
| CSolverTargetInterface | The interface to be provided by targets for non-linear equation solver |
| CSpectralPropagator | Spectral propagtor in time. Refer to documentation of file spectralprop.h for math detail |
| CSpectralPropagatorGaussLobatto | |
| CSpinlock | Spinlock using pthread spinlock operations |
| CSplit | Dummy class a la Intel TBB used to distinguish splitting constructor |
| CSRConf | |
| CStack | A simple, fixed-size, stack |
| CSteepestDescent | Unconstrained minimization via steepest descent |
| CStrongOrthogonalityProjector | SO projector class |
| CSubspace | The SubspaceK class is a container class holding previous orbitals and residuals |
| CSubspaceK | The SubspaceK class is a container class holding previous orbitals and residuals |
| CSystolicMatrixAlgorithm | Base class for parallel algorithms that employ a systolic loop to generate all row pairs in parallel |
| CTaggedKey | Key object that uses a tag to differentiate keys |
| CTaskAttributes | Contains attributes of a task |
| CTaskFn | Wrap a callable object and its arguments into a task function |
| CTaskFunction | |
| CTaskInterface | All world tasks must be derived from this public interface |
| CTaskMemfun | |
| CTaskThreadEnv | Used to pass info about thread environment into users task |
| CTDA | The TDA class: computes TDA and CIS calculations |
| CTDA_DFT | |
| CTDA_TIMER | Strucutre for TIMER |
| CTensorArgs | TensorArgs holds the arguments for creating a LowRankTensor |
| CTensorException | Tensor is intended to throw only TensorExceptions |
| CTensorIterator | |
| CTensorResultType | TensorResultType<L,R>::type is the type of (L op R) where op is nominally multiplication |
| CTensorTrain | |
| CTensorTypeData | Traits class to specify support of numeric types |
| CTensorTypeFromId | This provides the reverse mapping from integer id to type name |
| CThread | Simplified thread wrapper to hide pthread complexity |
| CThreadBase | Simplified thread wrapper to hide pthread complexity |
| CThreadPool | A singleton pool of threads for dynamic execution of tasks |
| CThreadPoolThread | ThreadPool thread object |
| CTimer | |
| Ctrajectory | |
| Ctrue_op | |
| CTwoElectronFactory | Factory for generating TwoElectronInterfaces |
| CTwoElectronInterface | Base class to compute the wavelet coefficients for an isotropic 2e-operator |
| CuniqueidT | |
| Cunperturbed_vxc | |
| CUnwrapReference | ReferenceWrapper type trait accessor |
| CUnwrapReference< detail::ReferenceWrapper< T > > | |
| CUnwrapReference< detail::ReferenceWrapper< T > const > | |
| CUnwrapReference< detail::ReferenceWrapper< T > const volatile > | |
| CUnwrapReference< detail::ReferenceWrapper< T > volatile > | |
| Cvecfunc | |
| CVector | A simple, fixed dimension Coordinate |
| CVectorOfFunctionsSpace | A vector space using MADNESS Vectors of MADNESS Functions |
| CVectorSpace | A vector space using MADNESS Vectors |
| CVextCosFunctor | A generic functor to compute external potential for TDDFT |
| CVLocalFunctor | |
| CVoid | A type you can return when you want to return void ... use "return None" |
| CWorld | A parallel world with full functionality wrapping an MPI communicator |
| CWorldAbsMaxOp | |
| CWorldAbsMinOp | |
| CWorldAmInterface | Implements AM interface |
| CWorldBitAndOp | |
| CWorldBitOrOp | |
| CWorldBitXorOp | |
| CWorldContainer | Makes a distributed container with specified attributes |
| CWorldContainerImpl | Internal implementation of distributed container to facilitate shallow copy |
| CWorldContainerIterator | Iterator for distributed container wraps the local iterator |
| CWorldDCDefaultPmap | Default process map is "random" using madness::hash(key) |
| CWorldDCPmapInterface | Interface to be provided by any process map |
| CWorldDCRedistributeInterface | |
| CWorldGopInterface | Provides collectives that interoperate with the AM and task interfaces |
| CWorldLogicAndOp | |
| CWorldLogicOrOp | |
| CWorldMaxOp | |
| CWorldMemInfo | Used to output memory statistics and control tracing, etc |
| CWorldMinOp | |
| CWorldMpiInterface | This class wraps/extends the MPI interface for World |
| CWorldMultOp | |
| CWorldObject | Implements most parts of a globally addressable object (via unique ID) |
| CWorldProfile | Singleton-like class for holding profiling data and functionality |
| CWorldProfileEntry | Used to store profiler info |
| CWorldProfileObj | |
| CWorldSumOp | |
| CWorldTaskQueue | Multi-threaded queue to manage and run tasks |
| CWSTAtomicBasisFunctor | |
| Cxc_functional | Class to compute the energy functional |
| Cxc_kernel | Class to compute terms of the kernel |
| Cxc_lda_potential | Compute the spin-restricted LDA potential using unaryop (only for the initial guess) |
| Cxc_potential | Class to compute terms of the potential |
| CXCfunctional | Simplified interface to XC functionals |
| CXCFunctionalLDA | |
| Cxfunction | The Root structure is needed by the TDA class |
| CXNonlinearSolver | Generalized version of NonlinearSolver not limited to a single madness function |
| ►Nmpfr | |
| Cconversion_overflow | |
| Cmpreal | |
| ►Nmu | Namespace for mathematical applications |
| ►NTest | Namespace for test cases |
| CParserTester | Test cases for unit testing |
| CParser | Mathematical expressions parser |
| CParserBase | Mathematical expressions parser (base parser engine) |
| CParserByteCode | Bytecode implementation of the Math Parser |
| CParserCallback | Encapsulation of prototypes for a numerical parser function |
| CParserComplex | Mathematical expressions parser |
| CParserError | Error class of the parser |
| CParserErrorMsg | A class that handles the error messages |
| CParserInt | Mathematical expressions parser |
| CParserStack | Parser stack implementation |
| CParserToken | Encapsulation of the data for a single formula token |
| CParserTokenReader | Token reader for the ParserBase class |
| CSTATIC_ASSERTION_FAILURE | Static type checks |
| CSTATIC_ASSERTION_FAILURE< true > | |
| ►NSafeMPI | |
| CException | SafeMPI exception object |
| CGroup | |
| CIntracomm | Wrapper around MPI_Comm. Has a shallow copy constructor; use Create(Get_group()) for deep copy |
| CRequest | |
| CStatus | |
| ►Nstd | |
| ►Ntr1 | |
| ►Ngtest_internal | |
| CAddRef | |
| CAddRef< T & > | |
| CByRef | |
| CByRef< T & > | |
| CGet | |
| CGet< 0 > | |
| CGet< 1 > | |
| CGet< 2 > | |
| CGet< 3 > | |
| CGet< 4 > | |
| CGet< 5 > | |
| CGet< 6 > | |
| CGet< 7 > | |
| CGet< 8 > | |
| CGet< 9 > | |
| CSameSizeTuplePrefixComparator | |
| CSameSizeTuplePrefixComparator< 0, 0 > | |
| CSameSizeTuplePrefixComparator< k, k > | |
| CTupleElement | |
| CTupleElement< true, 0, GTEST_10_TUPLE_(T) > | |
| CTupleElement< true, 1, GTEST_10_TUPLE_(T) > | |
| CTupleElement< true, 2, GTEST_10_TUPLE_(T) > | |
| CTupleElement< true, 3, GTEST_10_TUPLE_(T) > | |
| CTupleElement< true, 4, GTEST_10_TUPLE_(T) > | |
| CTupleElement< true, 5, GTEST_10_TUPLE_(T) > | |
| CTupleElement< true, 6, GTEST_10_TUPLE_(T) > | |
| CTupleElement< true, 7, GTEST_10_TUPLE_(T) > | |
| CTupleElement< true, 8, GTEST_10_TUPLE_(T) > | |
| CTupleElement< true, 9, GTEST_10_TUPLE_(T) > | |
| Ctuple | |
| Ctuple<> | |
| Ctuple_element | |
| Ctuple_size | |
| Ctuple_size< GTEST_0_TUPLE_(T) > | |
| Ctuple_size< GTEST_10_TUPLE_(T) > | |
| Ctuple_size< GTEST_1_TUPLE_(T) > | |
| Ctuple_size< GTEST_2_TUPLE_(T) > | |
| Ctuple_size< GTEST_3_TUPLE_(T) > | |
| Ctuple_size< GTEST_4_TUPLE_(T) > | |
| Ctuple_size< GTEST_5_TUPLE_(T) > | |
| Ctuple_size< GTEST_6_TUPLE_(T) > | |
| Ctuple_size< GTEST_7_TUPLE_(T) > | |
| Ctuple_size< GTEST_8_TUPLE_(T) > | |
| Ctuple_size< GTEST_9_TUPLE_(T) > | |
| ►Ntesting | |
| ►Ninternal | |
| CAddReference | |
| CAddReference< T & > | |
| CAssertHelper | |
| Cbool_constant | |
| CCompileAssert | |
| CCompileAssertTypesEqual | |
| CCompileAssertTypesEqual< T, T > | |
| CConstCharPtr | |
| CDefaultGlobalTestPartResultReporter | |
| CDefaultPerThreadTestPartResultReporter | |
| CEnableIf | |
| CEnableIf< true > | |
| CEqHelper | |
| CEqHelper< true > | |
| CFilePath | |
| CFloatingPoint | |
| CFormatForComparison | |
| CFormatForComparison< ToPrint[N], OtherOperand > | |
| CGTestFlagSaver | |
| CGTestLog | |
| CGTestMutexLock | |
| CHasNewFatalFailureHelper | |
| CImplicitlyConvertible | |
| Cis_pointer | |
| Cis_pointer< T * > | |
| CIsAProtocolMessage | |
| CIteratorTraits | |
| CIteratorTraits< const T * > | |
| CIteratorTraits< T * > | |
| Clinked_ptr | |
| Clinked_ptr_internal | |
| CMutex | |
| CNativeArray | |
| COsStackTraceGetter | |
| COsStackTraceGetterInterface | |
| CRandom | |
| CRE | |
| CRemoveConst | |
| CRemoveConst< const T > | |
| CRemoveConst< const T[N]> | |
| CRemoveReference | |
| CRemoveReference< T & > | |
| Cscoped_ptr | |
| CScopedTrace | |
| CSingleFailureChecker | |
| CStaticAssertTypeEqHelper | |
| CStaticAssertTypeEqHelper< T, T > | |
| CString | |
| CTestFactoryBase | |
| CTestFactoryImpl | |
| CTestPropertyKeyIs | |
| CTestResultAccessor | |
| CThreadLocal | |
| CTraceInfo | |
| CTypeIdHelper | |
| CTypeWithSize | |
| CTypeWithSize< 4 > | |
| CTypeWithSize< 8 > | |
| CUnitTestImpl | |
| CUnitTestOptions | |
| CUniversalPrinter | |
| CUniversalPrinter< T & > | |
| CUniversalPrinter< T[N]> | |
| CUniversalTersePrinter | |
| CUniversalTersePrinter< char * > | |
| CUniversalTersePrinter< const char * > | |
| CUniversalTersePrinter< T & > | |
| CUniversalTersePrinter< T[N]> | |
| CUniversalTersePrinter< wchar_t * > | |
| ►Ninternal2 | |
| CTypeWithoutFormatter | |
| CTypeWithoutFormatter< T, kConvertibleToInteger > | |
| CTypeWithoutFormatter< T, kProtobuf > | |
| CAssertionResult | |
| CEmptyTestEventListener | |
| CEnvironment | |
| CMessage | |
| CScopedFakeTestPartResultReporter | |
| CTest | |
| CTestCase | |
| CTestEventListener | |
| CTestEventListeners | |
| CTestInfo | |
| CTestPartResult | |
| CTestPartResultArray | |
| CTestPartResultReporterInterface | |
| CTestProperty | |
| CTestResult | |
| CUnitTest | |
| CAtom | Abstract Atom class |
| CAtomCore | |
| CAtomicBasis | Represents multiple shells of contracted gaussians on a single center |
| CAtomicBasisFunction | Used to represent one basis function from a shell on a specific center |
| ►CAtomicBasisSet | Contracted Gaussian basis |
| CAnalysisSorter | |
| CAtomicData | |
| CbaseWF | |
| CBoundWF | |
| CCFFT | |
| CContractedGaussianShell | Represents a single shell of contracted, Cartesian, Gaussian primitives |
| CCoreOrbital | |
| CCorePotential | Represents a core potential |
| CCorePotentialManager | |
| CCoupledPurturbation | |
| CCubicInterpolationTable | An class for 1-D data interpolation based on cubic polynomials |
| CDBF | A d basis function |
| CDFTSolventSolver | |
| CDirichletCondIntOp | The operator needed for solving for 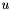 with GMRES with GMRES |
| CDirichletLBCost | |
| CEFieldOperator | |
| CEnvelopedPulse | |
| CExampleClass1 | Every class should be documented (this is the brief line) |
| CExampleClass2 | Brief documentation of second example class in group |
| CExpikr | |
| CFrequencyHandler | |
| CFrequencyIncident | |
| Cgauss_function | Gauss_function structure is needed to mimic noise |
| CGaussian | |
| CGaussianBF | Abstract Gaussian basis function. Must be overriden to specify angular momenta (s, p, d, etc.) |
| CHydrogen | Hydrogen atom |
| CInputParameters | |
| CKPoint | |
| CLevelPmap | |
| CMolecularEntity | |
| CMolecularMaskBase | |
| CMolecularNuclearChargeDensityFunctor | |
| CMolecularSurface | |
| CMolecularSystem | |
| CMolecularVolumeComplementMask | |
| CMolecularVolumeExponentialSwitch | Switches between positive values Vint and Vext with special log derivative |
| CMolecularVolumeExponentialSwitchLogGrad | Returns the requested component of the derivative of the log of MolecularVolumeExponentialSwitch |
| CMolecularVolumeExponentialSwitchReciprocal | Computes the reciprocal of MolecularVolumeExponentialSwitch |
| CMolecularVolumeMask | |
| CMolecularVolumeMaskGrad | |
| CMolecule | |
| CMPI_Status | |
| CNO_DEFAULTS | Disables default copy constructor and assignment operators |
| Cnoise | Try to create noise using random gauss functions |
| COutputWriter | |
| CParserHandler | |
| CPBF | A p basis function |
| CPhiK | |
| CPhikl | |
| CPointGroup | |
| CPurturbationOperator | |
| Creal_op | |
| Croot | POD holding excitation energy and response vector for a single excitation |
| CSBF | An s basis function |
| ►CScatteringWF | |
| CMemberFuncPtr | |
| CSCF | |
| CSCFParameters | |
| CScreenSolventPotential | |
| CSVPEColloidSolver | |
| CTimeIncident | |
| CTipMolecule | Setup the tip-molecule problem |
| CTiXmlAttribute | |
| CTiXmlAttributeSet | |
| CTiXmlBase | |
| CTiXmlComment | |
| CTiXmlCursor | |
| CTiXmlDeclaration | |
| CTiXmlDocument | |
| CTiXmlElement | |
| CTiXmlHandle | |
| CTiXmlNode | |
| CTiXmlOutStream | |
| CTiXmlPrinter | |
| CTiXmlString | |
| CTiXmlText | |
| CTiXmlUnknown | |
| CTiXmlVisitor | |
| CWF | |
| CWSTFunctional | |
| Cxc_functional | Class to compute the energy functional |
| Cxc_lda_potential | Compute the spin-restricted LDA potential using unaryop (only for the initial guess) |
| Cxc_potential | Class to compute terms of the potential |
| CXCfunctional | Simplified interface to XC functionals |
| CYl0 |
![\[ z = frac{x}{1 - y^2} \]](form_43.png)
 1.8.8
1.8.8